5. Bruker TruLive3D Experiment settings
This page will guide through the steps to set-up a basic experiment.
Make sure you performed the calibration of the system before starting with your sample.
- Finding your sample
- Install the sample in the sample holder and into the sample chamber. Hold it at one end and push the other end against the end of the brackets then press the other end into place.
- You can use the space mouse to move the dish to its position above the detection objective.
- Click on Dashboard in LuxBundle.
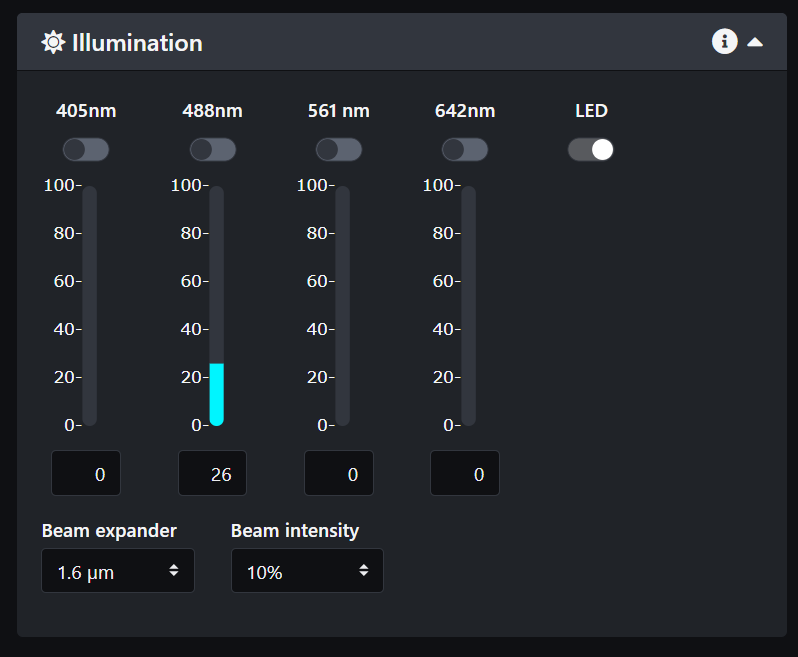
- In the illumination panel, turn on the LED light.
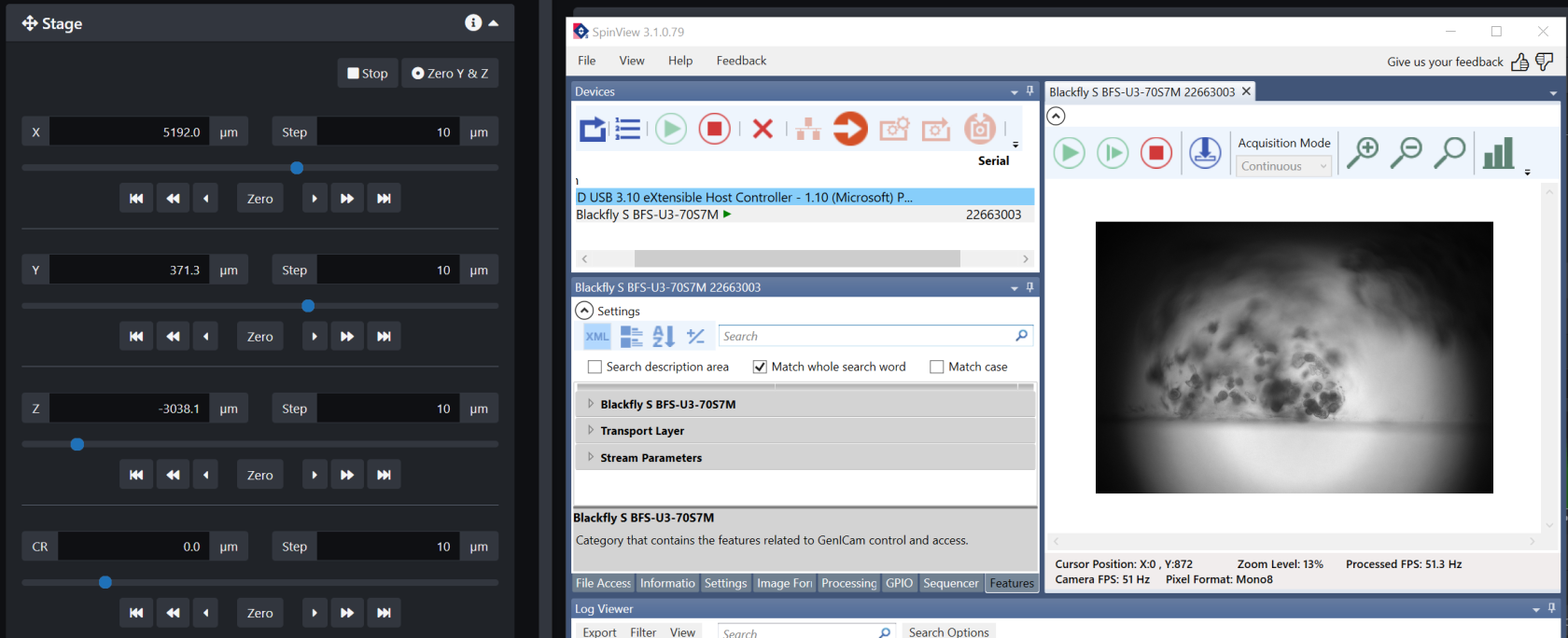
- Open the SpinView software and select the Blackfly camera then click the green arrow. There is a small camera in the system that looks through one of the illumination objectives.
- Click on the image area and scroll out to see the entire field of view
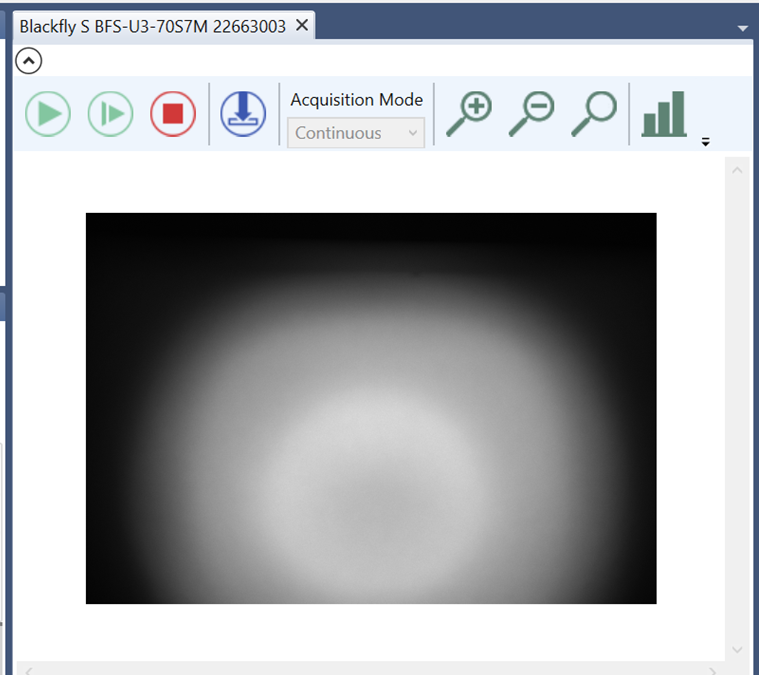
- Go back to LuxBundle and use the stage control buttons to move the sample until it appears in the field of view of the Blackfly camera.
- If your sample is already above the detection objective, move the Z stage to lower it.
- If it is not above the detection objective, move the X stage as well.
- Move the Y stage to bring it in focus and confirm that what you are looking at looks like what you expect and is not an artifact.
NOTE: This is not the focus of the detection objective! It brings the sample in focus of the illumination objective the Blackfly camera is looking through. This step is for identification only.
- Now that you know the sample is in the right location, turn off the Blackfly camera by clicking on the red square and turn off the LED light in LuxBundle.
- Select the laser lines you need for your sample.
- Select the emission filters for each camera. If you are not sure which filters to select for your sample please refer to page A. Bruker TruLive3D emission filters and fluorophores spectrum and try to pick a pair of filters that prevents any overlap in emission.
- Connect both cameras and start live
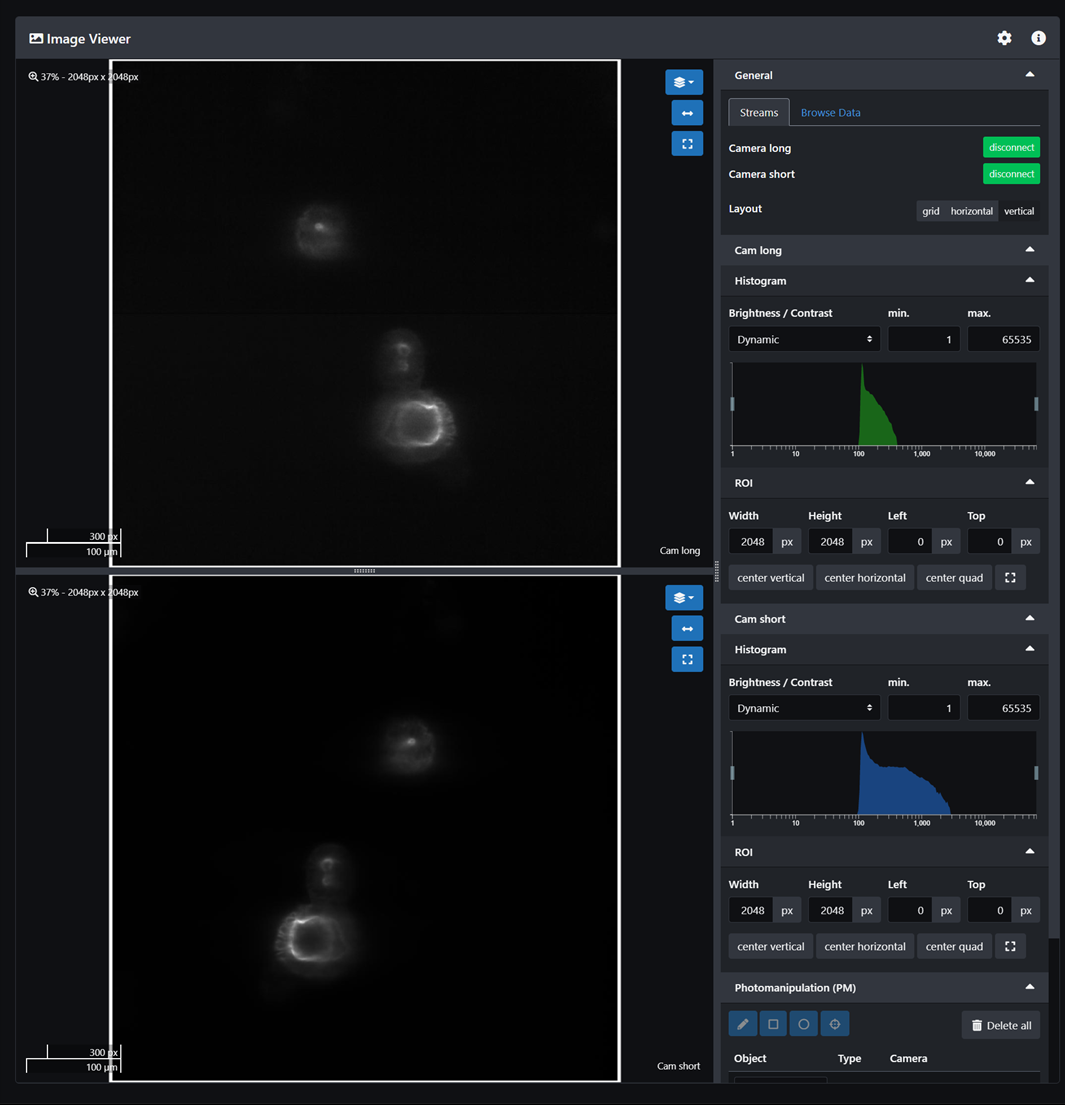
- Adjust the camera exposure and select either line mode or area mode.
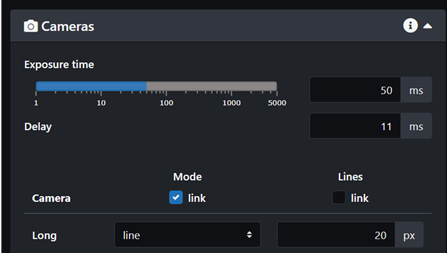
- Find a location of interest in the sample using the stage controls. Adjusting the Correction Ring (CR) might sharpen your image. You should start exploring your sample in Z to have an idea of the dimensions of the z-stack you will want to image.
2. Setting up an experiment
You have many possibilities to set-up an experiment with the Bruker TruLive3D. You can record a single z-stack at several time points, multiple z-stacks at several time points and you can also acquire bigger areas than the field of view by tiling the images together.
You can acquire data in a single channel with one or multiple laser lines on and simultaneously acquire images for two different emission channels, or you can create several channels for each laser line.
NOTE: if you only need one camera to acquire your images make sure you turn off the other one in the camera panel (unlink them and turn one off) otherwise both cameras will capture images which doubles the size of your data.
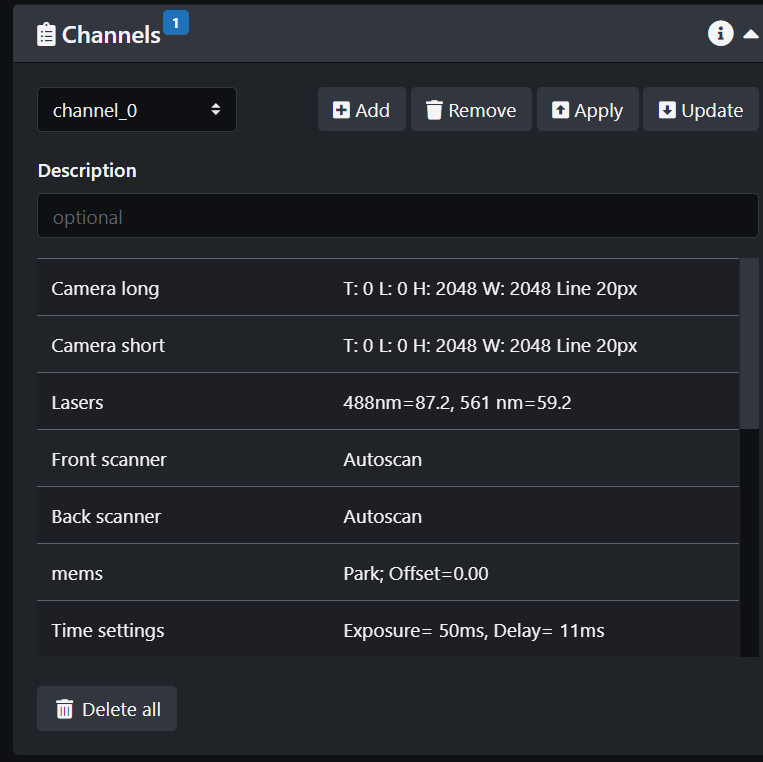
- Create a channel. This will have all the current savings on the screen: laser lines and power, beam expander setting, detection filters, camera settings. If you change a setting, remember to update the channel.
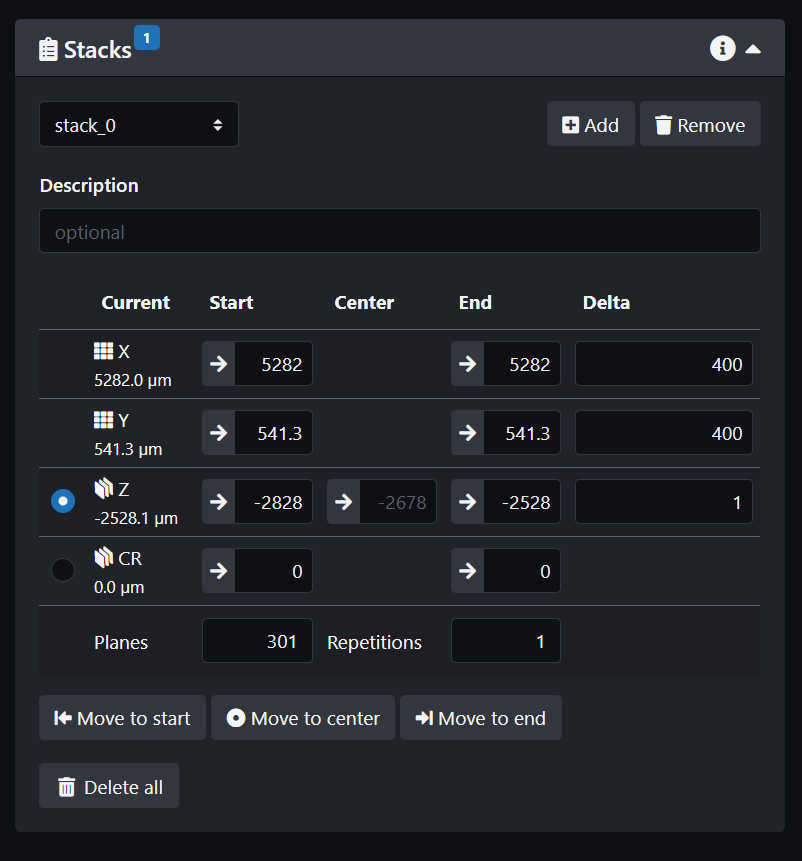
- Create a z-stack. Move the Z stage until you find the stack you want. At the bottom of the stack, click on the arrow in the z-stack parameter to update the value read on the stage control panel. Then go to the top of the stack and repeat the process. To visualize the data, the z-stack panel has buttons to go directly to the middle, bottom or top of the stack.
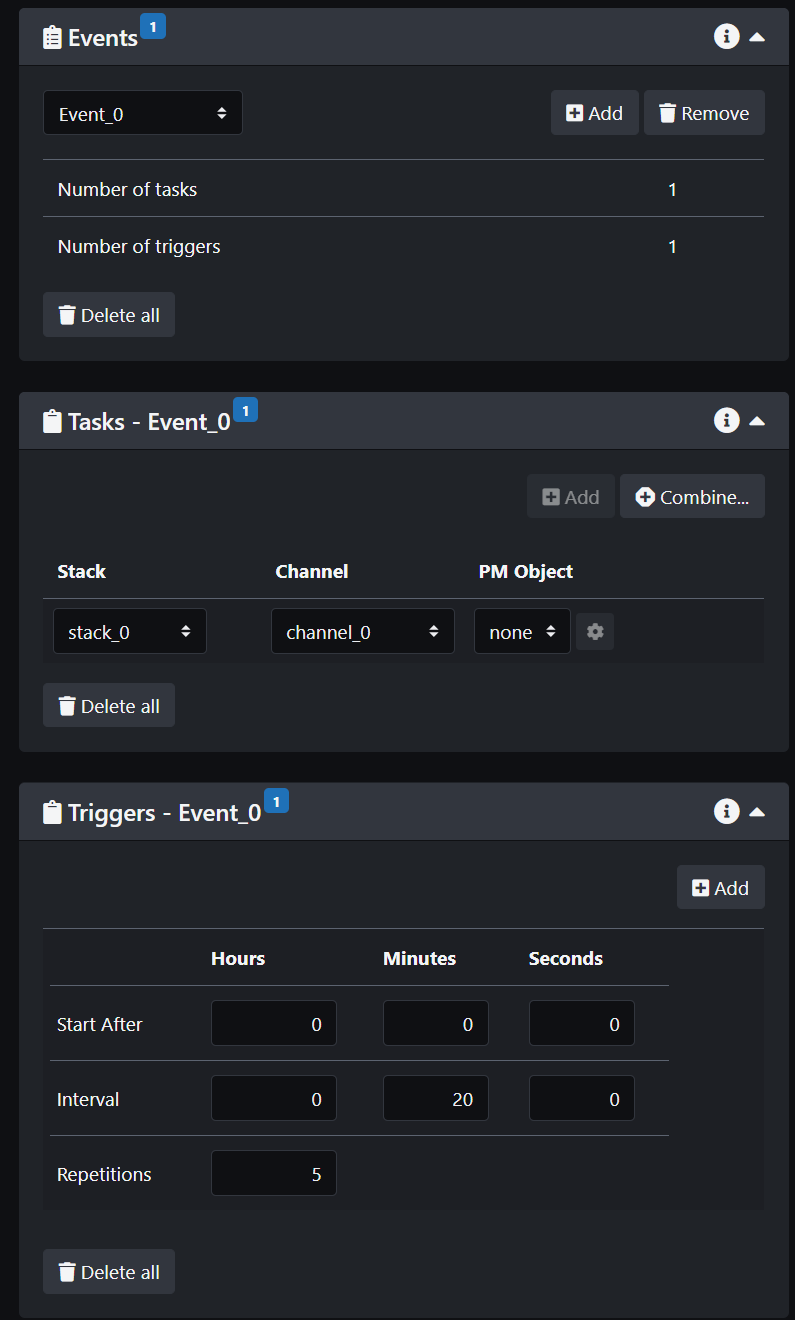
- Create an event. This will open two other panels with tasks and trigger. Set up your task(s) with the channel(s) you want and the z-stack(s) you want. You can have the same channel for all the different z-stacks or different channels with different z-stacks.
- Set-up your trigger. This defines the parameters for the timelapse.
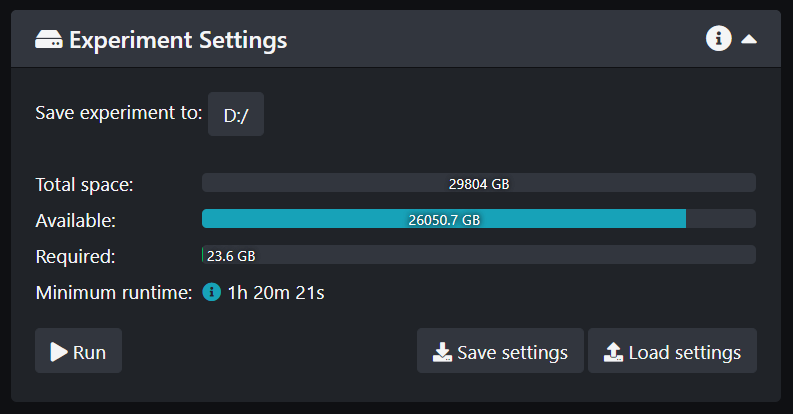
- Select where you want to save your data. If you are saving your data locally, create a folder for your Group in the Disk D and select this folder. If you are saving directly on your server (preferred method) select it as a location. To be able to save directly on your server you must ask the Beckman Center admin to set it up. Follow the procedure taped on the wall to name your data: Lab_Initials_Date_DataDetails
- Click Run when everything is ready! You can also save your settings for future use. Make sure you save those settings in your folder.
- When you experiment is done please refer to the Data management page to follow the procedure to transfer and delete it.
3. Visualize your data
- Click on image viewer
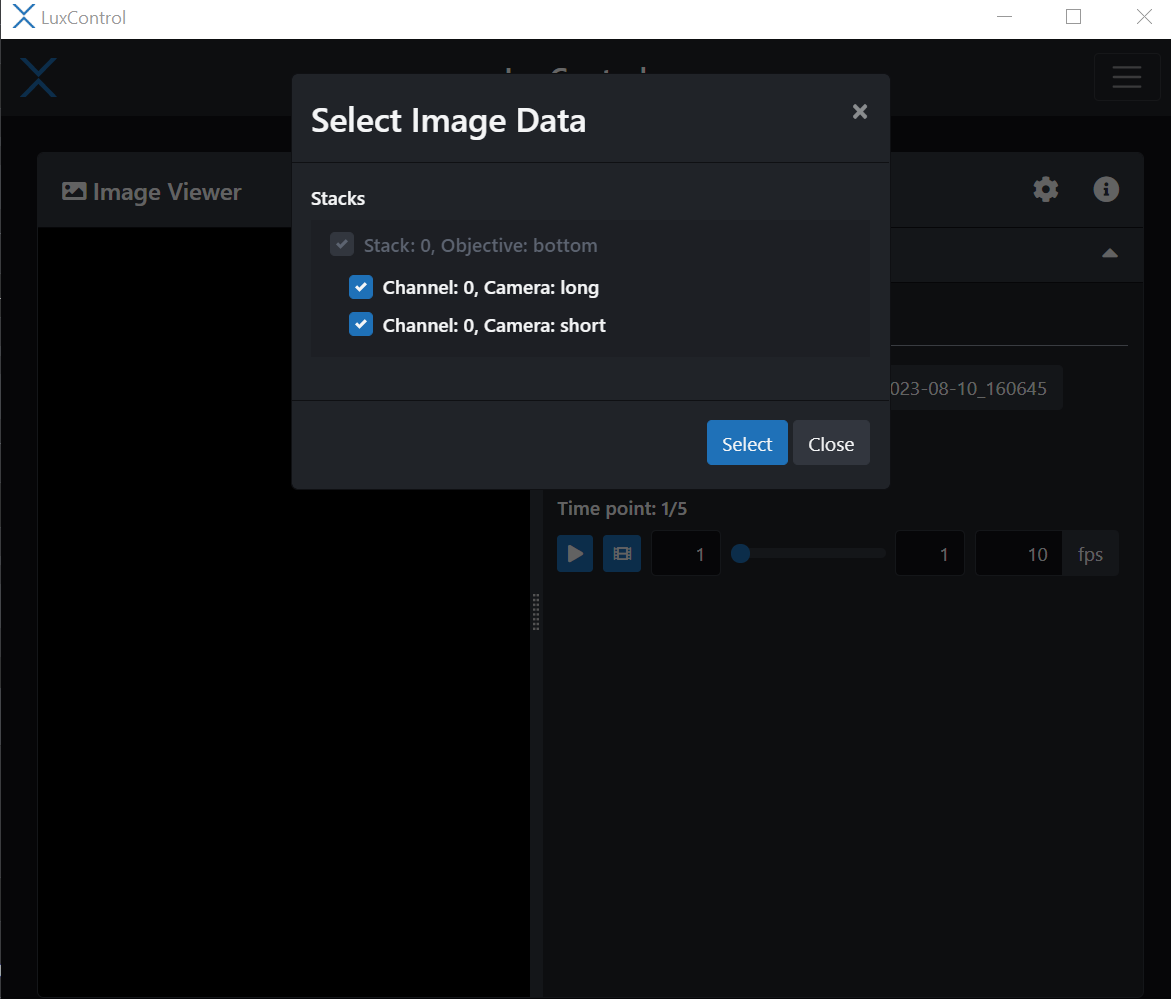
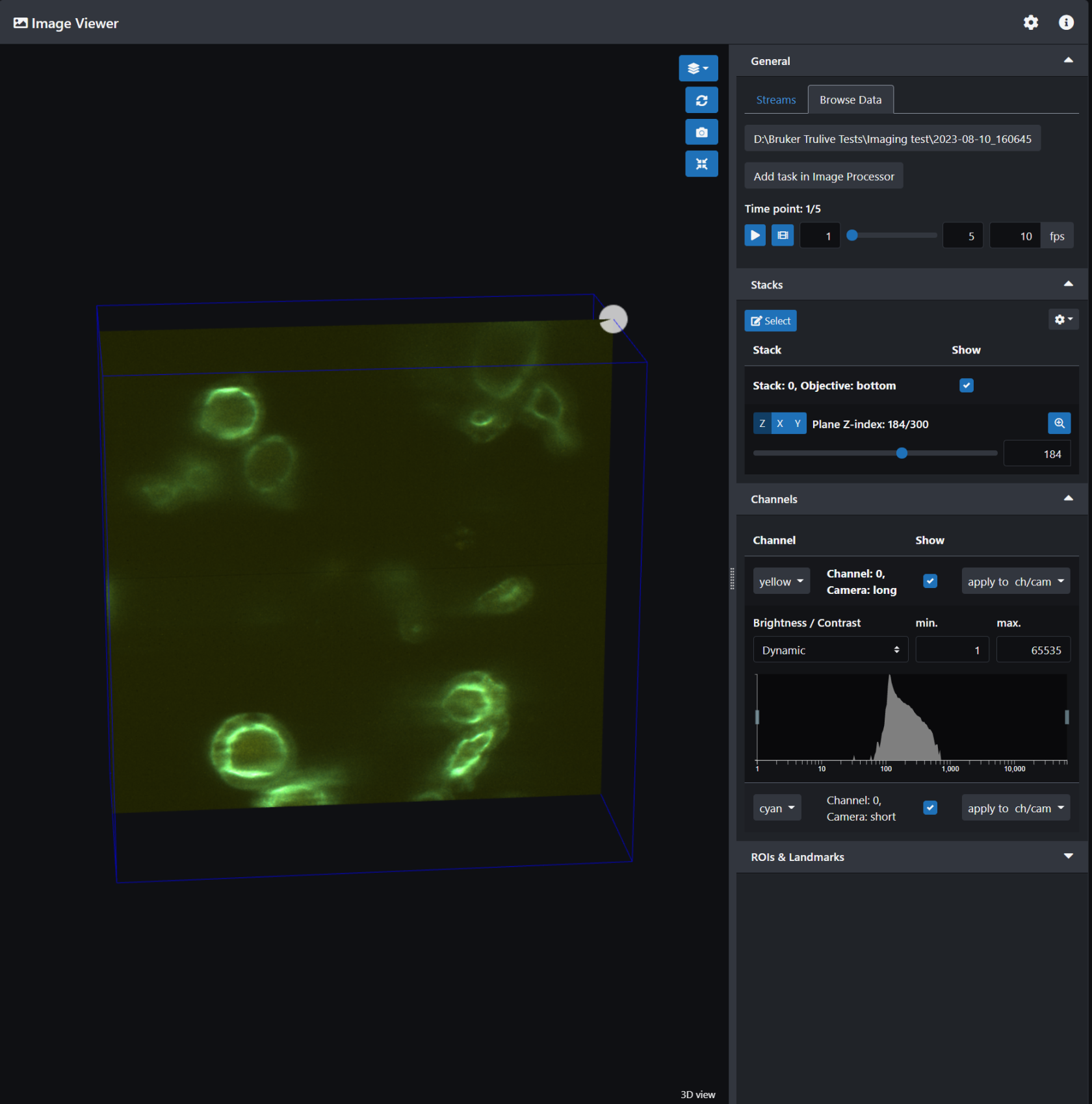
The image viewer in LuxBundle allows you to visualize the data frame by frame. It is not a 3D viewer, unlike Imaris viewer, but can give you an idea of what happened during your experiment at each time point. You can also play a movie of the z-stack. When you open the image viewer you can select the image data you want.
Finally, LuxBundle has an image processor function that allows you to do a few things to process your data.
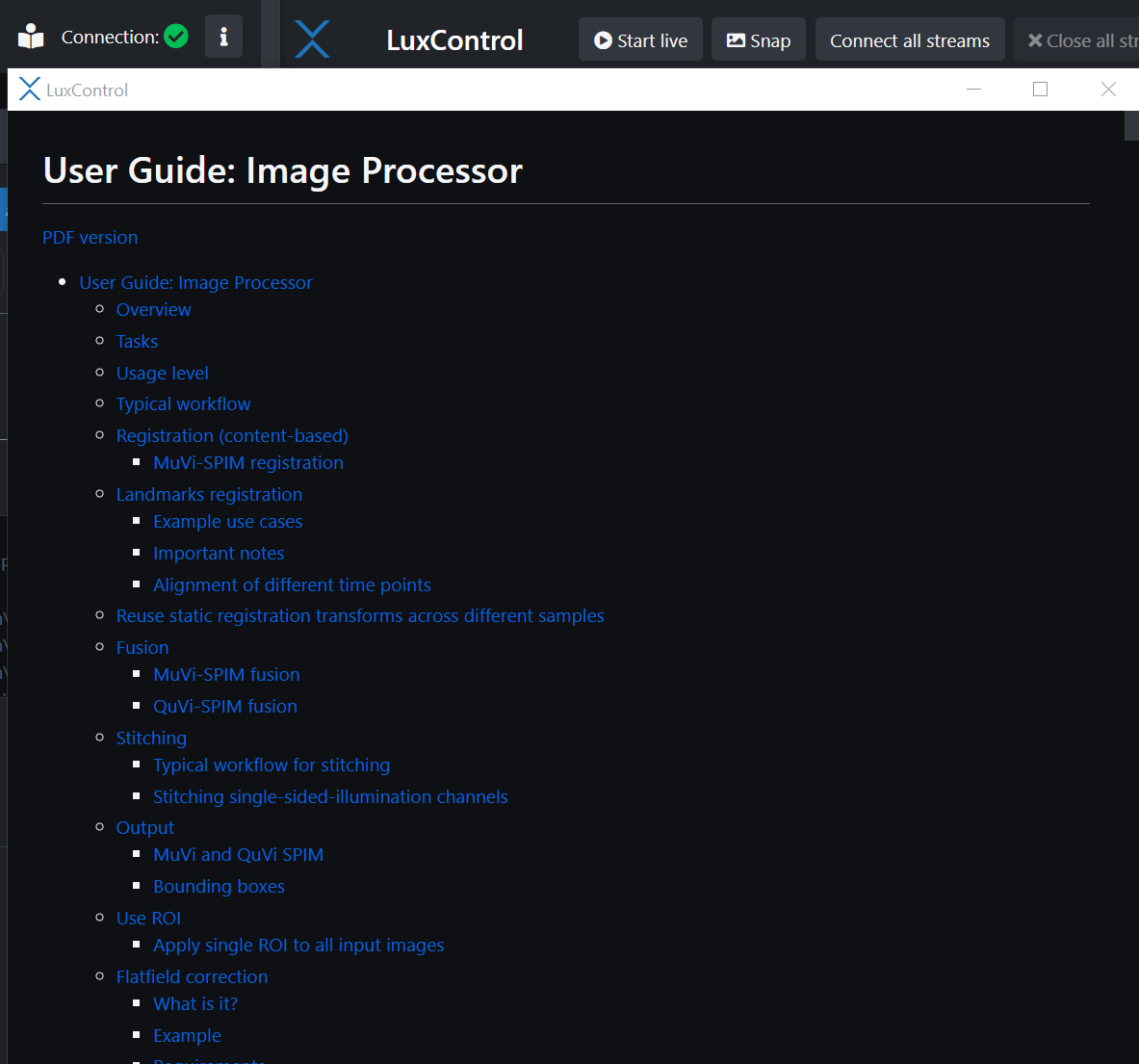
For a complete overview of the capabilities of the Image Processor, please read the User Guide located in the little book icon on the top right corner of the LuxControl window.
When your data is acquired it automatically creates an imaris header file along the Fiji Big Data viewer header files. However, depending on the size of your data set loading the images in imaris can take a very long time.
The image processor enables you to process your data stack by stack and create separate imaris header files.
Here, we will guide you through the steps to perform this task. For more information and potential debugging please refer to the User Guide in LuxBundle. You can also download the PDF version to read offline.
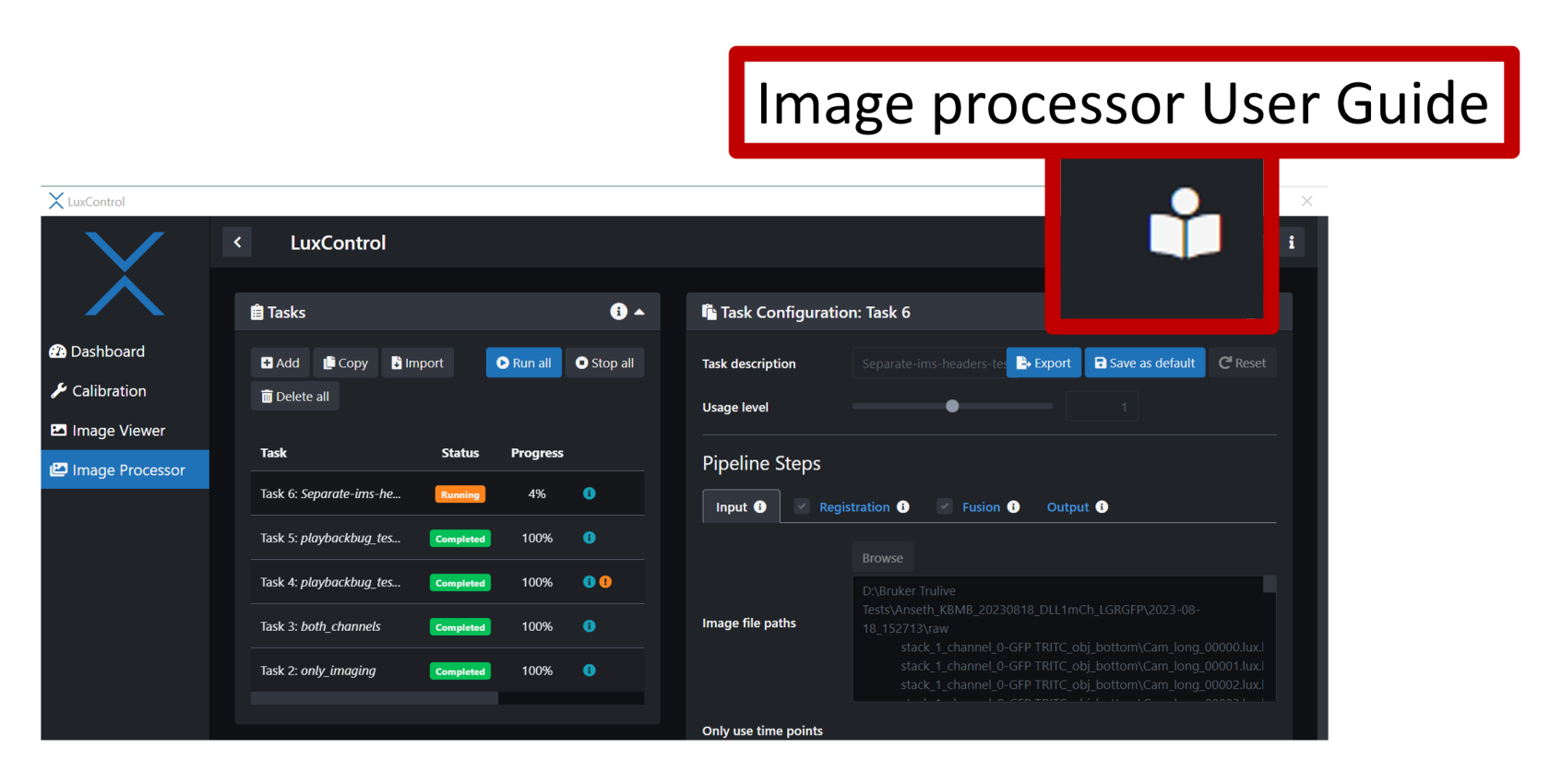
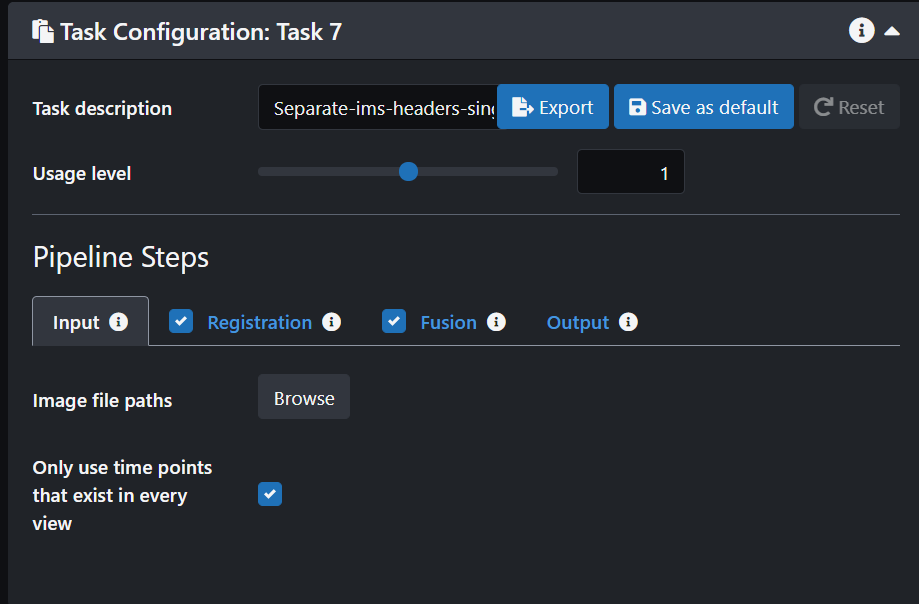
- Add a new Task in the Tasks panel
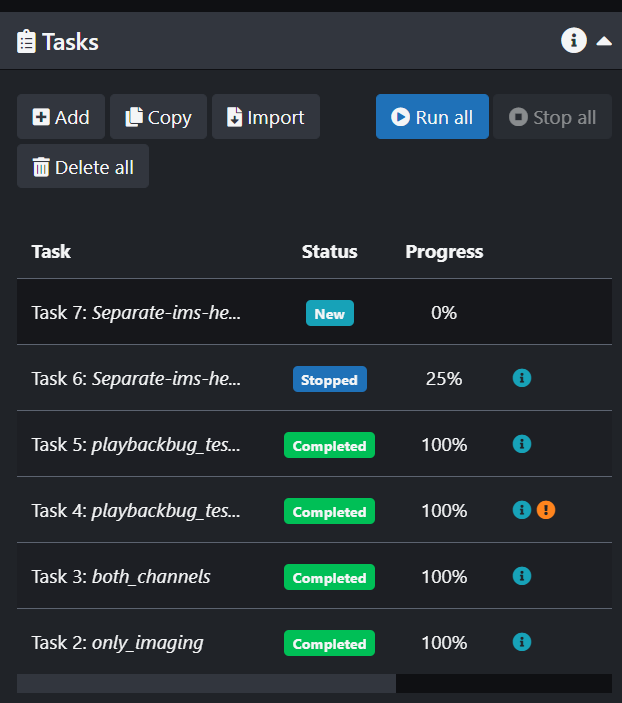
- In the Task Configuration panel Name your new task and then click on Input.
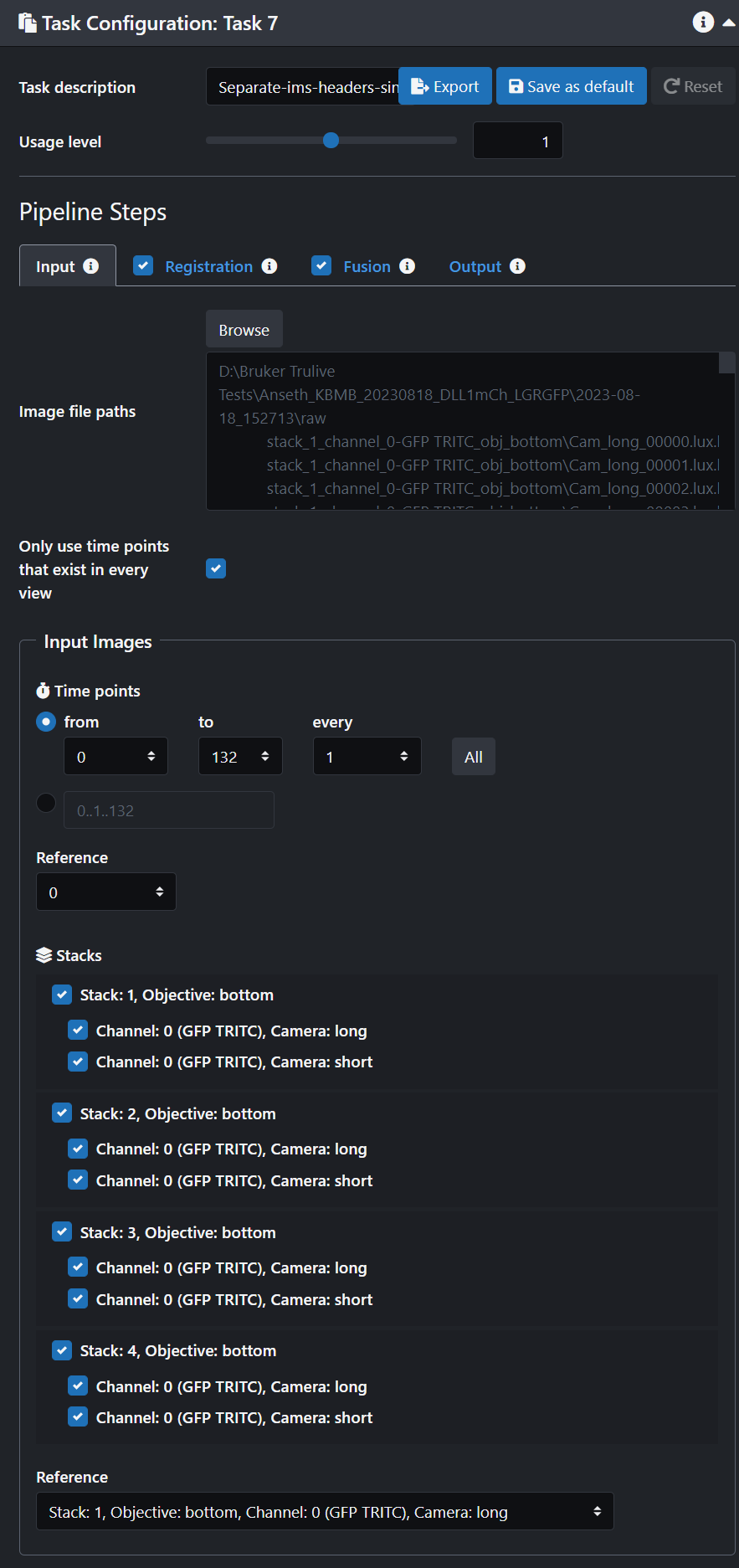
- Browse through our data to select the image folder that you would like to process.
- If you only want to process a few stacks then de-select the ones you don't want.
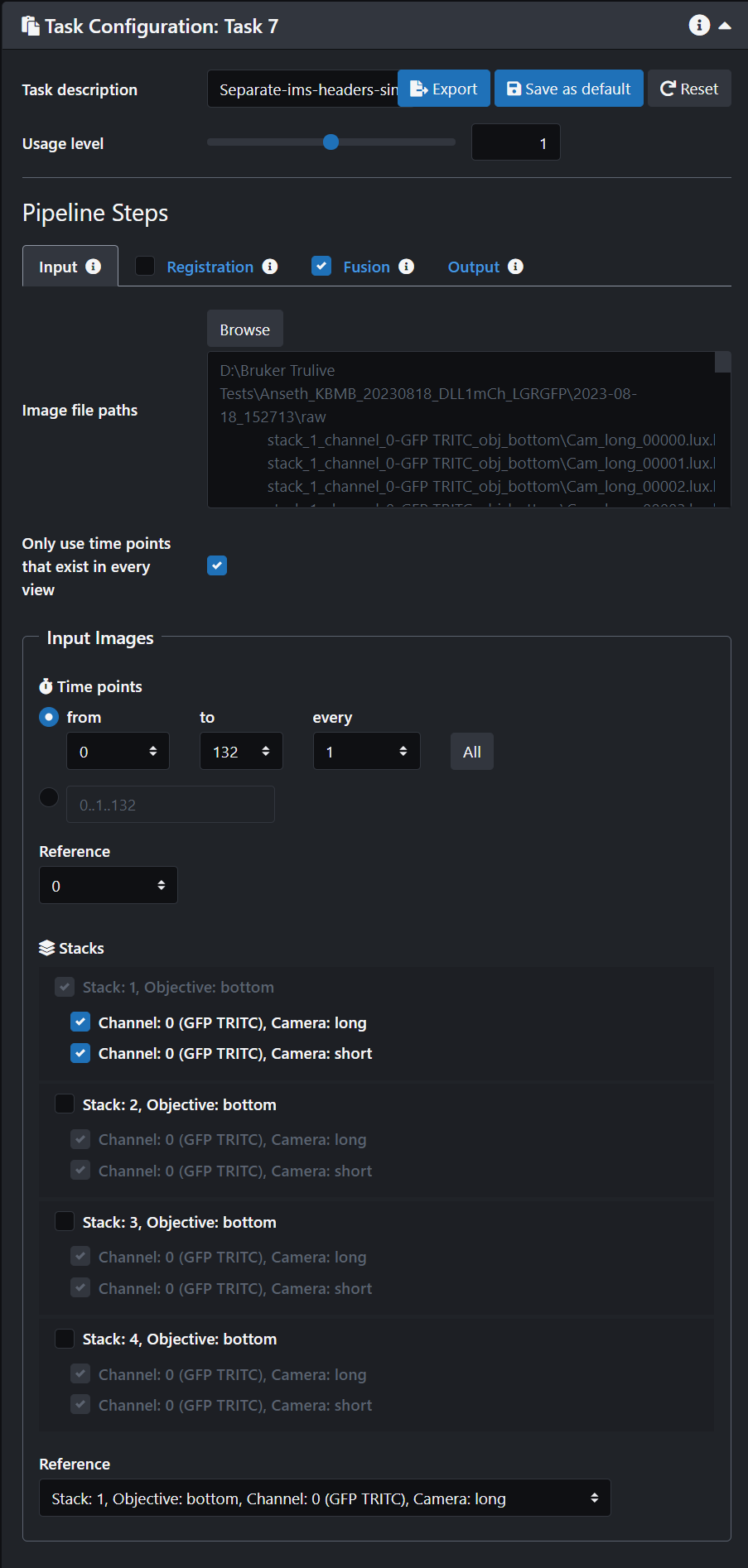
- Click on the Registration tab and turn it off.
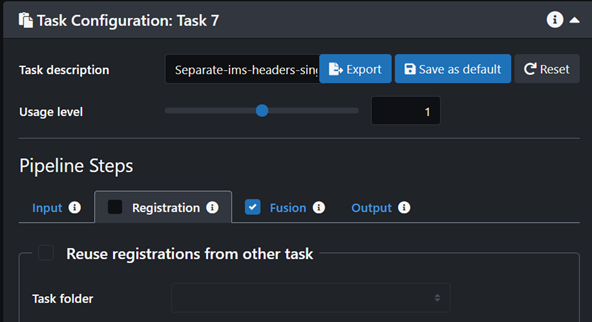
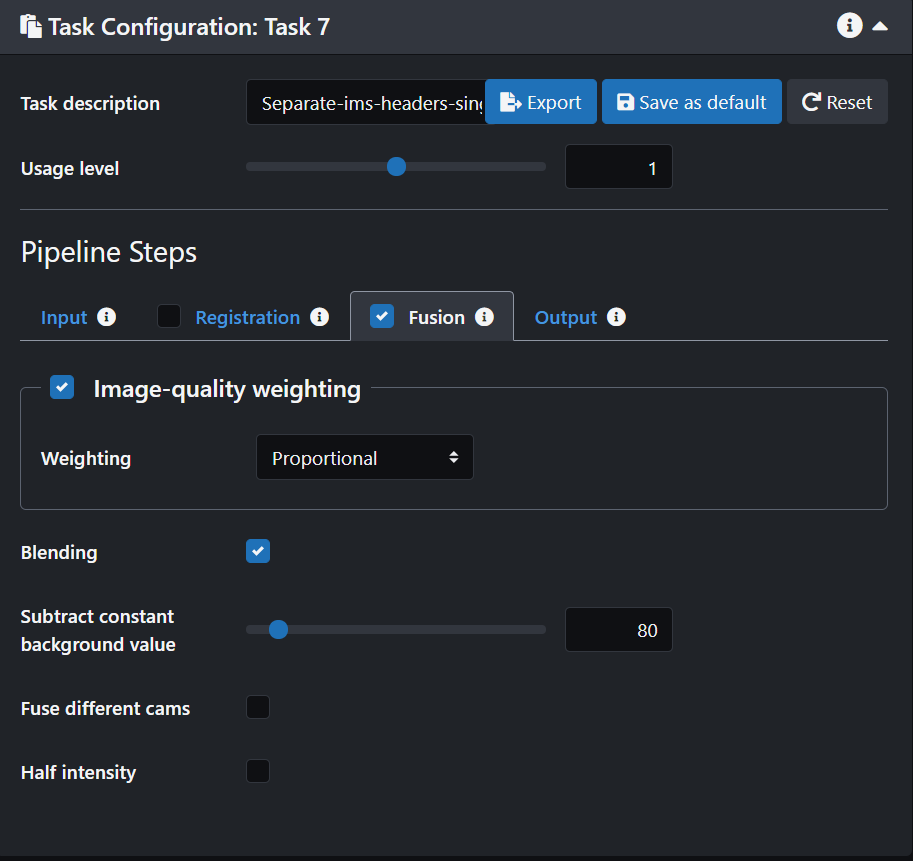
- Click on the Fusion tab and turn it on
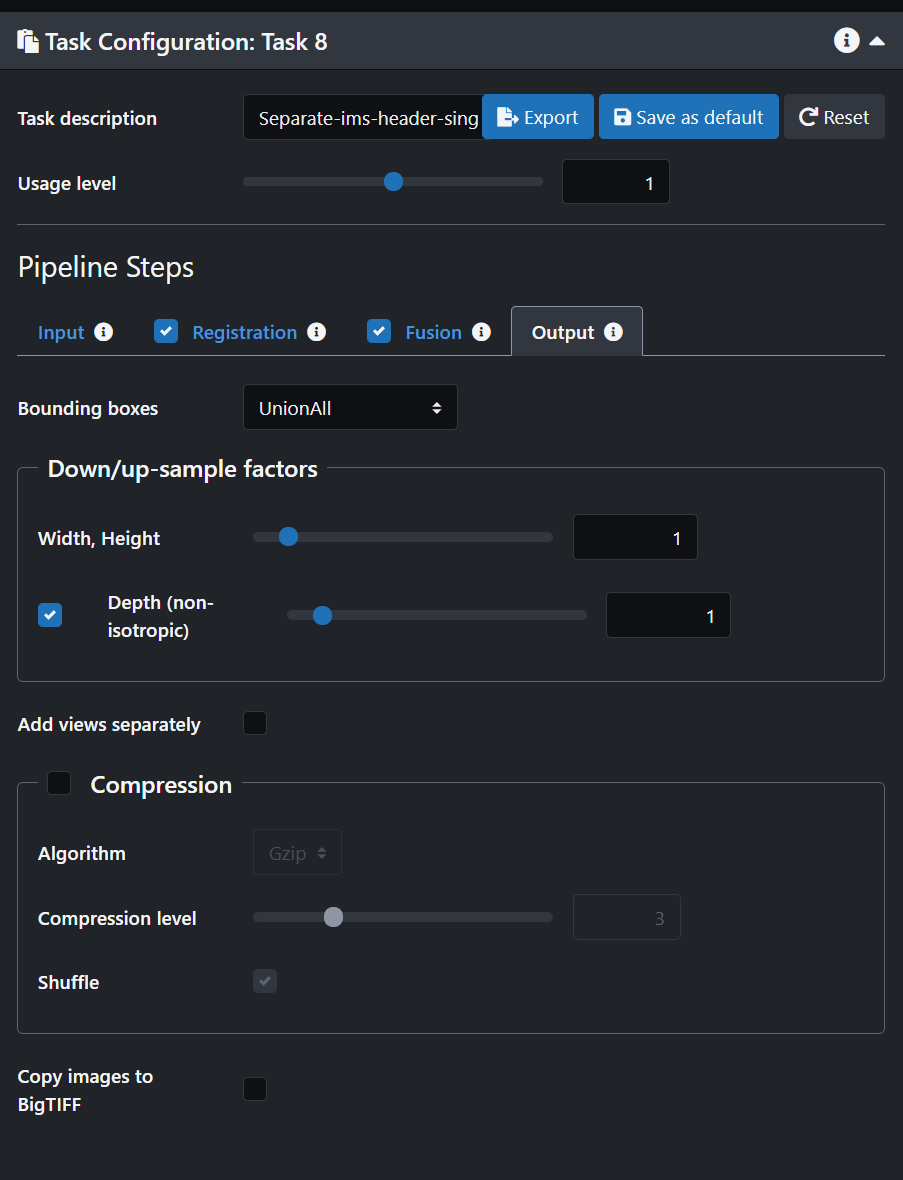
- Click on the Output tab and select "Separate" on the "Bounding boxes" option. Select "Depth (non isotropic)"
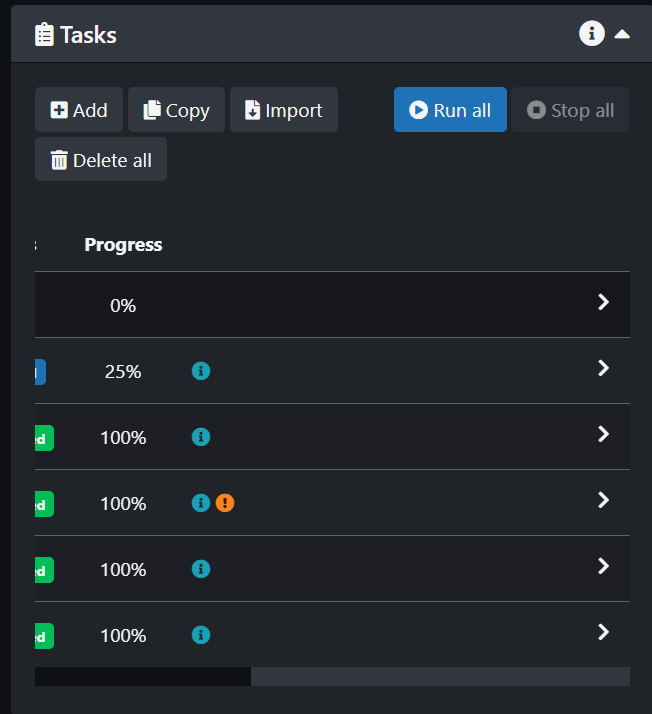
- The next step is to run the Task. Go back to the Tasks panel and scroll to the right then click on the arrow and click run. You can also stop or delete the task in this menu.
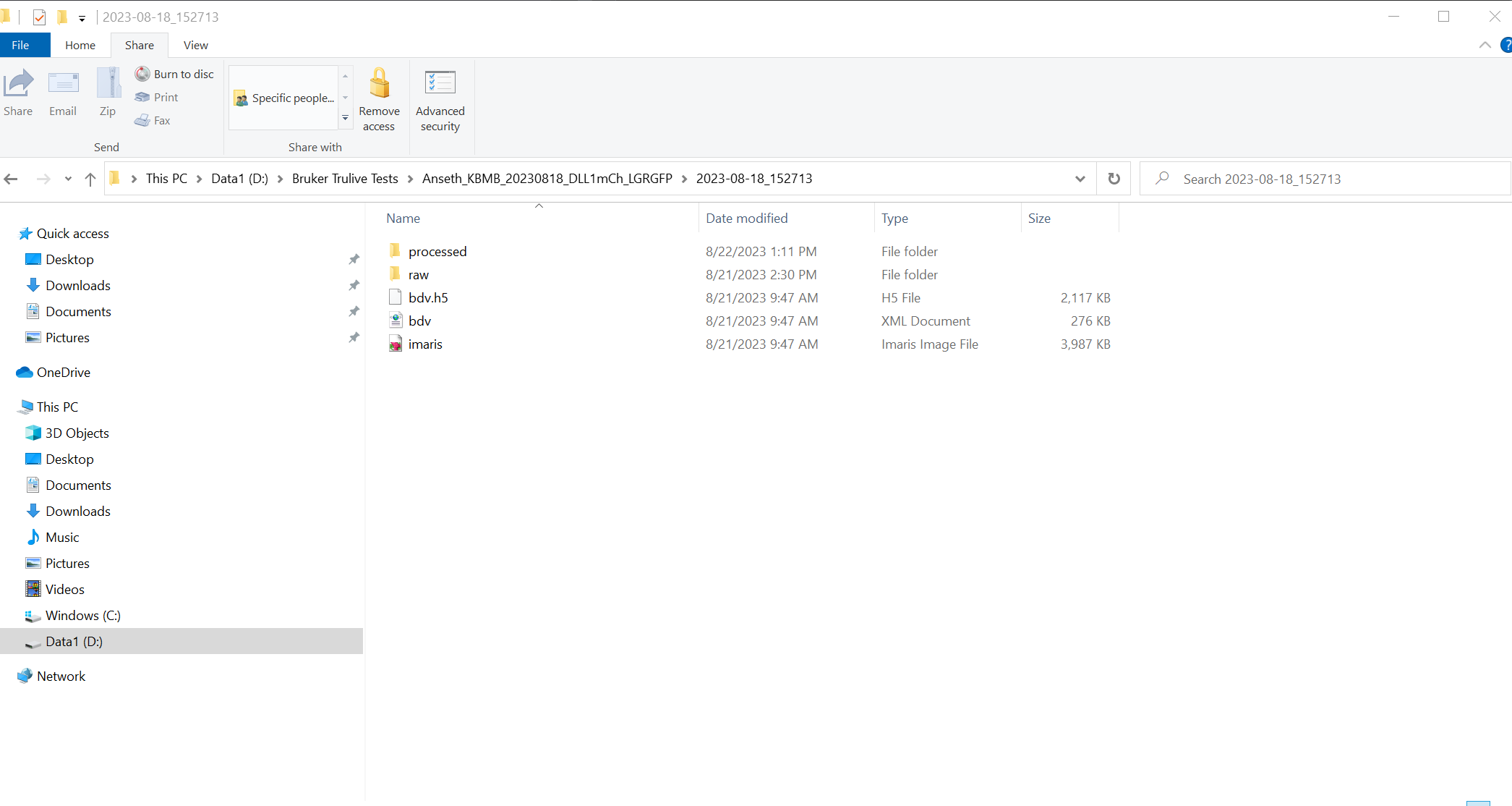
- When you go to your image folder you will now see a "processed" folder that contains the separate imaris headers and the task configuration parameters as a JSON file.