Machine learning segmentation
This section explains how to use the ML segmentation tool in Imaris.
- In the Surpass view, click on the Surfaces icon in the left panel to create a new Surface.

- A wizard dialog box will appear on the bottom left of the screen.

- On this page:
- Unselect Classify Surfaces – classification is used to distinguish between different objects, e.g., different cell types, and we won’t need it here.
- Unselect Object-Object Statistics – these statistics can be used, for example, to filter objects by distance to its nearest neighbors. Disabling these statistics will reduce the computational time and power needed for the segmentation. You can always re-enable this after segmentation.
- Select Start creation with Slicer view
- Click the blue right arrow to move to the next page.
- Set the following options:

- Set Source Channel to Channel 1 – W1-DAPI.
- Check the Smooth option – Smoothing is generally a good idea to avoid jagged surfaces. The default value for the Surfaces Detail is equivalent to a 2-pixel width, which we’ll use for this example.
- Select Machine Learning Segmentation. Leave the All Channels box unchecked.
- Click on the blue right arrow to move to the next screen.
- The next screen shows the machine learning training panel. You will spend most of the time on this screen as you teach the machine how to segment your images.

- The Background class labels pixels that should be considered background.
- The Foreground class labels pixels that are in your objects of interest (cell nuclei in this case).
- To label the image:
- Select the class for the pixels you want to label.
- Shift + Left click to start labeling.
- Ctrl + Mouse wheel can be used to change the size of the brush stroke. Alternatively, you can zoom in/out using the mouse wheel to draw more specific areas.
- If you make a mistake, you can click on the Delete Last button to undo the last annotation. Note there is no way to select a specific annotation to delete – you can only undo one previous action at a time.
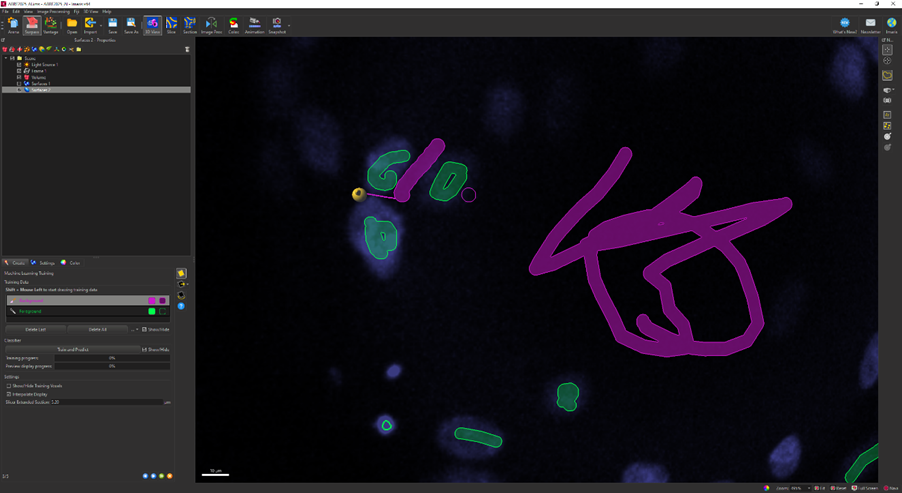
- Under Settings, make sure Interpolate Display is checked. This option will cause the Slicer view to show an interpolated volume, rather than a single slice. Likewise, your annotations will be over a volume, rather than a single slice.
- The volume used can be changed by the value in the Slicer Extended Section.
- The volume used can be changed by the value in the Slicer Extended Section.
- Annotating the image is where the art of using this tool comes in. Here is some general advice:
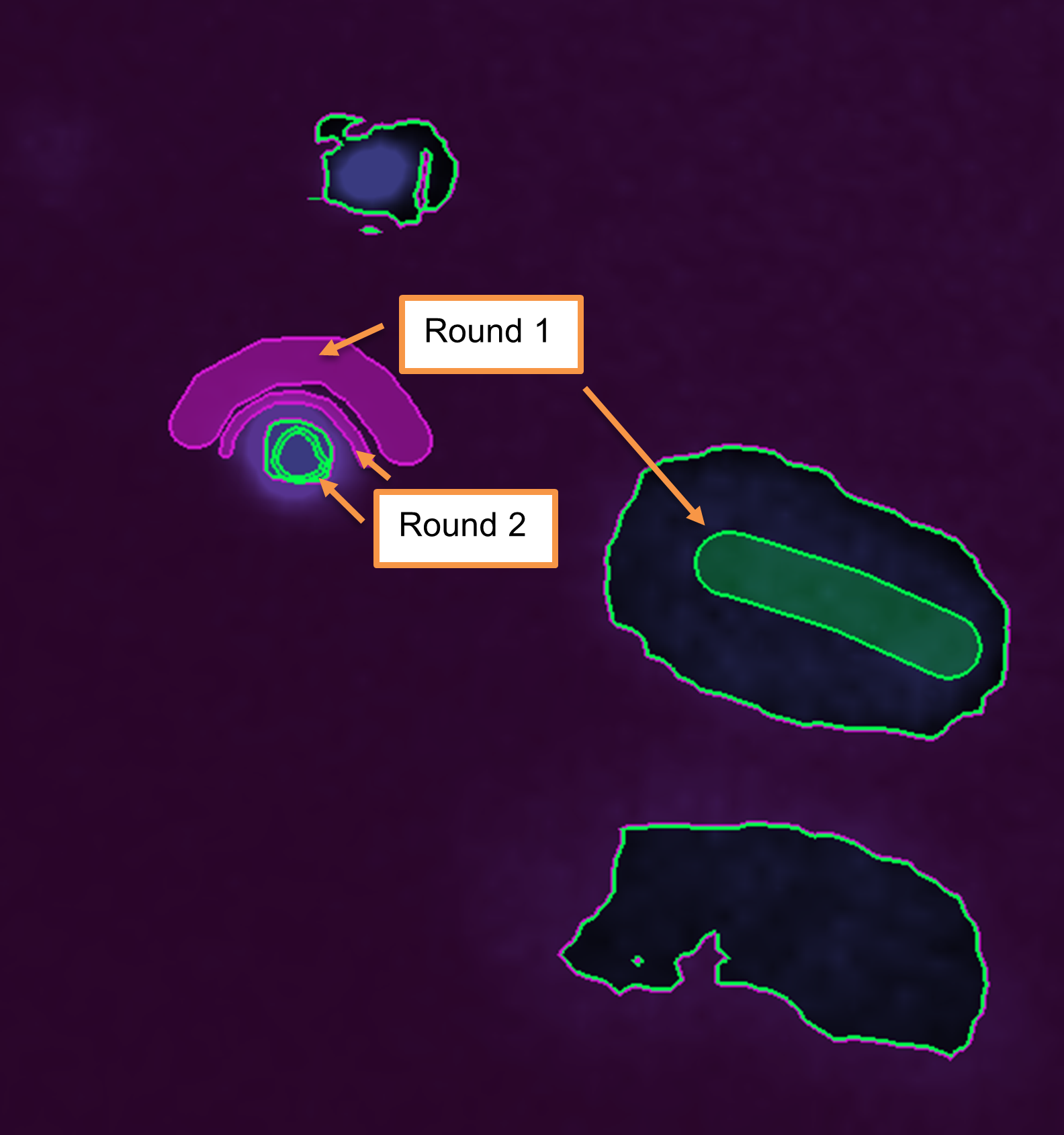
- Rather than labeling every single pixel, it is better to iterate the results over several rounds of training.
- In the first round, use some rough strokes to select pixels within a few cells and the background. You don’t need to color in a whole cell.
- If there are touching cells, it is generally a good idea to select a few background pixels that are between the cells.
- When you have a few annotations, click on Train and Predict to allow the machine to learn from your annotations.

- It is likely that the algorithm will get some areas wrong, so continue to refine and repeat the training process. Don’t forget to change the viewed slice to check other planes as well.
- When you are happy with the results (or you get tired of refining the mask), click on the blue right arrow to move to the next screen.
Important: If the results do not seem to be improving, or they seem to be getting worse, it is likely that the model is becoming overtrained. In this case, it can be helpful to delete a few annotations or even to Delete All and start again.
- You can select the option to Split Touching Objects, but let’s leave this for now and click on the green double right arrow to complete the segmentation.
Reusing your models
Training a model is hard work and you will likely want to save this model to reuse on other images. Note that the model will likely only work on images from the same dataset (i.e., collected on the same microscope, with the same settings, likely on the same batch of cells).
To save the model:
- After exiting the wizard, click on the Wizard tab in the left panel.
- Click on the Store Parameters for Batch… button.
- In the resulting dialog box, you can name your model. It is recommended that you select the boxes to save the model both as a Favorite Creation Parameter and in the Arena.
To reuse the model:
- As before, open your image and create a new Surface.
- On the first page of the wizard, select the saved model under Favorite Creation Parameters.
- You can then click through the wizard until you reach the Machine Learning Training panel (usually the third screen of the wizard). Imaris will use your previously trained model as a starting point to label the images.
- You can then refine as needed on the new image.

No Comments