Specifications and primary uses of the OPM
Single Objective Light Sheet Microscope
The Single Objective Light Sheet Microscope is also referred to as Oblique Plane Microscope or OPM. It is a custom design Light Sheet microscope that enables high resolution and can be used with standard coverslipped samples. There is virtually no sacrifice in numerical aperture (NA) or loss in resolution - unlike most light sheet microscopes.
The OPM therefore presents a strict improvement over standard confocal microscopy. It has the sectioning capability and reduced photo-toxicity of light sheet microscopes while preserving image quality.
The animation below illustrates how it works.
High_NA_single-objective_light-sheet_scan.mp4
For more technical information on the OPM design, check this open source paper.
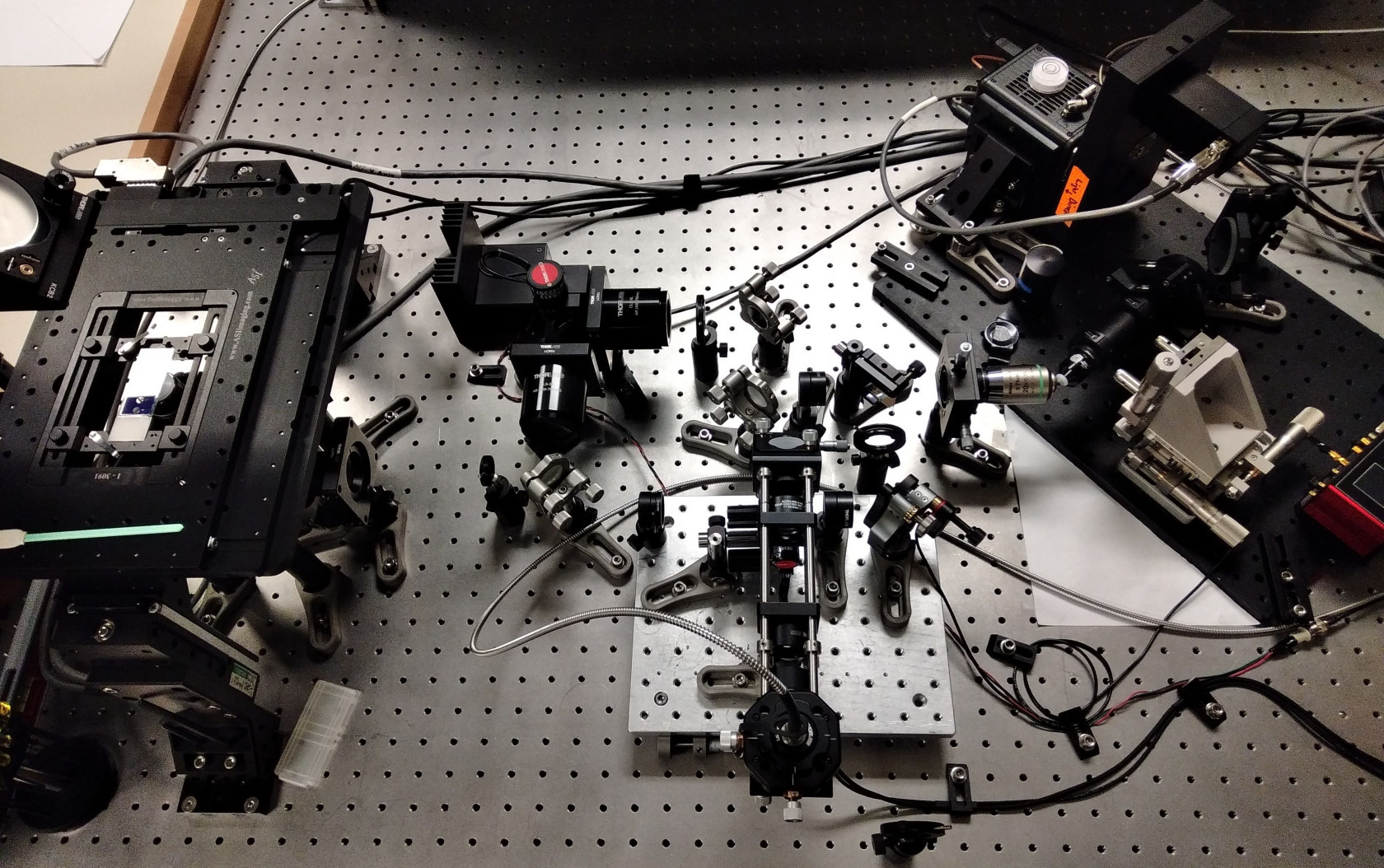
The Beckman Center Single Objective Light Sheet Microscope
- Primary objective: Olympus 20X 1.0NA water dipping
- Secondary objective: Nikon 20X 0.75NA
- Tertiary objective: AMS-AGY V1.0 "Snouty"
- Hamamatsu Fusion sCMOS camera
- ASI FTP2000 XYZ stage
- 4 Channels: 405, 488, 561, 640
The sample stage has several sample holders for slides, round dishes and multi-well plates.
Primary uses
The size range of the samples is roughly from single cells to the bottom layer of small drosophila or zebra fish embryos and small organoids. Typically the OPM is used for 1 to 3 cells deep into the sample.
While this limits the variety of sample that can be imaged on the OPM, the flexibility of sample preparation is a considerable advantage. The fact that is uses the same dishes as other standard commercial microscope allows for straightforward comparison of images between a confocal microscope and the single objective light sheet microscope.
Examples
Some imaging examples using the OPM technology by The Dean Lab at UT South-Western, Dallas Texas:
- Biological imaging of clathrin-mediated endocytosis, vimentin and membrane dynamics
- Natural killer cell mediated cytotoxicity
- Imaging in biological microchannels - microtubules and nuclear shielding
- High-speed imaging of calcium transduction and cytoplasmic flows
- Simultaneous volumetric imaging and optogenetic stimulation
- Large field of view imaging of cortical neurons and ventral furrow formation
- Tissue-scale imaging:
- "In addition to the rapid laser scan/descan illumination geometry, OPM is also compatible with a sample scanning acquisition format that is essentially field of view unlimited. Indeed, by combining scan optimized equipment with fully automated fluidic handling, it is possible to image ~1 cm2 of a thin tissue in less than 45 min per color and perform biochemistry, such as sequential multiplexed labeling. To demonstrate this, we imaged an entire 30-micron thick slice of coronal mouse brain tissue (Figure 9A, Video 15) labeled with the nuclear marker DAPI. Within these data, even small features like nucleoli are clearly resolved from both lateral and axial viewing perspectives throughout the entire ~6×8 mm tissue slice (Figure 9B and C). Likewise, we also imaged a ~ 4×14 mm slice of 12-micron thick human lung tissue labeled for nuclei, angiotension-converting enzyme 2 (ACE2) mRNA, and surfactant protein C (SFTPC) protein (Figure 9D). Here, characteristic histological features, including bronchiole, alveoli and vasculature, are readily visible, albeit with molecular contrast and sub-cellular resolution (Figure 9E,F and G, and Video 16). Quantification of molecular expression within this tissue section provides spatial information on ~20,000 cells, and verifies our previous limited quantification of ACE2 expression in alveolar epithelial type II cells using confocal microscopy (Muus and Luecken, 2020). Indeed, because we were not sterically restricted by the orthogonal illumination and detection geometry (Figure 9—figure supplement 1), the lateral dimensions of this human lung specimen were 8- and 1.5-fold larger than those of the biggest sample imaged with lattice light-sheet microscopy (Gao et al., 2019). However, in the third dimension, lattice light-sheet microscopy has in principle a 6.7x larger reach (2 mm working distance of the typically employed NA 1.1/25X detection objective compared to 300 microns working distance of our primary objective). In practice, optical aberrations limit high-resolution light-sheet microscopy to depths of a few hundreds of microns, even for highly transparent samples. Furthermore, our approach is fully compatible with automated fluid exchange, which is increasingly important for projects like the Human Cell Atlas that necessitate iterative imaging approaches for spatial -omics of RNAs and proteins at the single-cell level throughout entire tissues (Chen et al., 2015)."
For further details and reference, the original article can be found here.